
Loss of habitat and human activities such as fishing and shipping pose a grave threat to wildlife but diseases driven by the smallest organisms in the ocean are a less understood side of marine conservation.
These diverse and abundant microbiome communities perform complex processes on skin and tissue of marine wildlife – and Flinders University scientists are breaking ground by understanding their role in an endangered shark species and to describe new marine microbes for the first time.

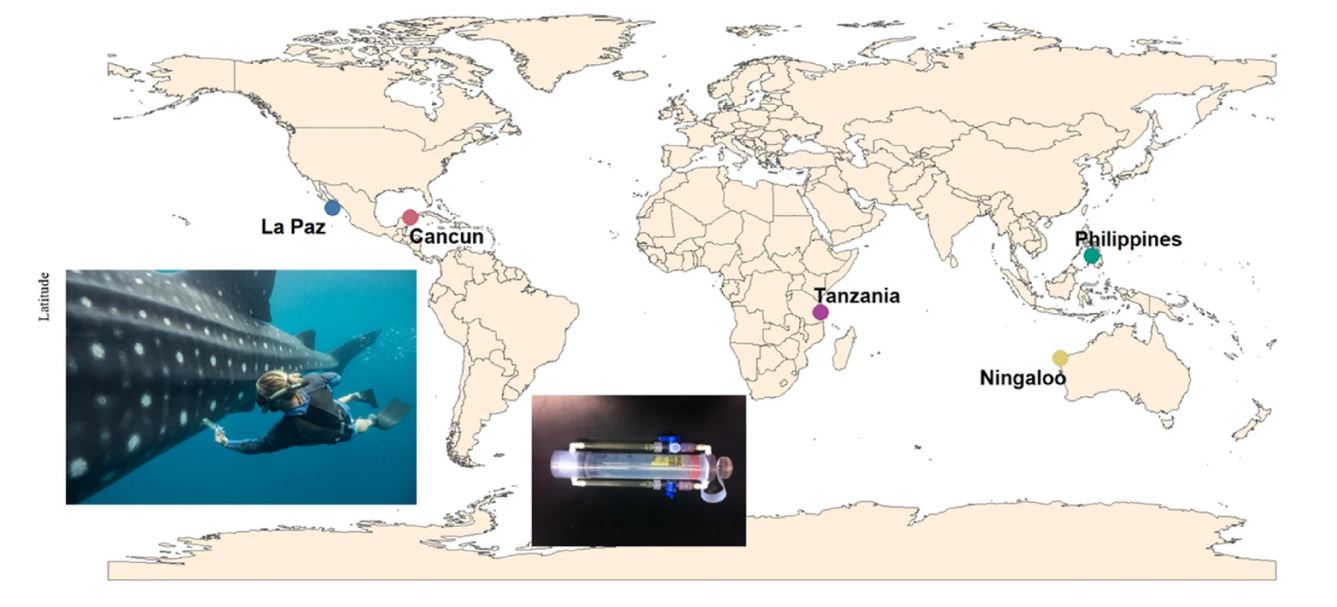
In a new article, scientists from around the world have collaborated to sample microbes on the skin surface of the world’s largest fish – the whale shark (Rhincodon typus) – at five of the most famous diving sites around the world, including Ningaloo Reef in Western Australia, Oslob in the Philippines, Mafia Island in Tanzania and La Paz and Cancún in Mexico.
With these docile filter-feeding sharks, the scientists from 12 international institutions collected microbial samples and then used cutting-edge genomic sequencing technology to describe the types of microbes on the skin surface of the whale sharks.
The study is the most extensive microbiome study to date of a wild marine animal of this physical size, involving 74 whale sharks in the three major ocean basins, and will form a baseline for future analysis and highlight how microbial species differ around the world, says Dr Michael Doane, a researcher from the Flinders Accelerator for Microbiome Exploration (FAME) group in South Australia.
“While microbial species differ across the world, they work together to form a balanced network that contributes to the health of the sharks.
“It is important to measure and analyse the distinct and diverse epidermal microbiome of the global whale shark populations to work towards understanding how the microbes affect the wellbeing and survival of this amazing animal.
“The characteristics of a balanced microbial community are not well described for any species, but especially for sharks, which form a vital link in ocean foodchains and ecosystems.
“The microbes form a complex network pattern on the skin surface, which is consistent across sharks from each location, revaling characteristics of what comprises a balanced or unbalanced microbiome.”
Mathematical modelling was used to investigate how the microbes interacted with one another to form a community or network.
In addition to revealing the emergent structure of the shark skin microbiome, the study also described 34 new species of microbes.

The whale sharks from Ningaloo had the highest number of novel microbial species, “suggesting we have lots to learn about the microbial organisms inhabiting animals in Australian waters,” adds Flinders University Professor Elizabeth Dinsdale, co-author of the new study in Scientific Reports.
Professor Dinsdale says the right balance of microbes are critical to the well-being and survival of the host with which they live.
“In humans, for example, skin microbes are always present, and when these communities are ‘healthy’, they go unnoticed; however, when they are unbalanced, they cause skin conditions such as dermatitis, or skin infections,” she says.
“As microbes are too small to see, it is important to gain an understanding of how these complex network patterns fit together for a healthy microbiome balance. Unbalanced interactions can lead to a loss of benefits for the host and can result in disease.
“We now have a baseline for the healthy microbiome of whale sharks, which can be used to monitor the health of the whale shark and we also developed a non-invasive method to identify the effectiveness of conservation strategies.”
Microscopic organisms conduct important processes including nitrogen and oxygen production in oceans, consuming organic matter and metabolising carbohydrates.
 The article, Emergent community architecture despite distinct diversity in the global whale shark (Rhincodon typus) epidermal microbiome (2023) by Michael P Doane, Michael B Reed, Jody McKerral, Laís Farias Oliveira Lima, Megan Morris, Asha Z. Goodman, Shaili Johri, Bhavya Papudeshi, Taylor Dillon, Abigail C Turnlund, Meredith Peterson, Maria Mora, Rafael de la Parra Venegas, Richard Pillans, Christoph A Rohner, Simon J Pierce, Christine G Legaspi, Gonzalo Araujo, Deni Ramirez-Macias, Robert A Edwards and Elizabeth A Dinsdale has been published in Scientific Reports. DOI: 10.1038/s41598-023-39184-5.
The article, Emergent community architecture despite distinct diversity in the global whale shark (Rhincodon typus) epidermal microbiome (2023) by Michael P Doane, Michael B Reed, Jody McKerral, Laís Farias Oliveira Lima, Megan Morris, Asha Z. Goodman, Shaili Johri, Bhavya Papudeshi, Taylor Dillon, Abigail C Turnlund, Meredith Peterson, Maria Mora, Rafael de la Parra Venegas, Richard Pillans, Christoph A Rohner, Simon J Pierce, Christine G Legaspi, Gonzalo Araujo, Deni Ramirez-Macias, Robert A Edwards and Elizabeth A Dinsdale has been published in Scientific Reports. DOI: 10.1038/s41598-023-39184-5.
Acknowledgements: We would like to thank Stephanie Lo and Ben Billings Global Shark Research and Conservation Fund. Field work in Tanzania was supported by Aqua-Firma, WWF Tanzania, the Shark Foundation, Waterlust and two private trusts. In the Philippines we thank the Tan-awan Oslob Sea Warden and Fishermen Association and permission from the Municipality of Oslob. Export of water samples from the Philippines were under export permit BFAR-7 n from the Bureau of Fisheries and Aquatic Resources. Sampling in Cancun and La Paz were under permit 03362 from the Secretaria de Medio Ambiennte Y Recursoso Naturales.
Animal handling and ethics were reviewed at San Diego State University through IACUC under permit APF #14-05-011D, APF #17-11-010D, APF # 18-05-007D. The raw metagenomic datasets generated for this project are available under BioProject accession PRJNA808622.

